Load Balancing Molecular Dynamics Simulation
Due: November 20
The purpose of this assignment is to analyze the benefits of load balancing on a system where the load changes and for an application where the application load also changes, i.e. containing both resource and application dynamics.
Write a MPI-based code for simulating molecular dynamics with the same overall structure as seen in the PPP class. You can make this as simple as you want, e.g. 1D instead of 2D decomposition, no diagonal movement etc. In order to calculate new positions and velocities of molecules, use the following Verlet Velocity integrator:
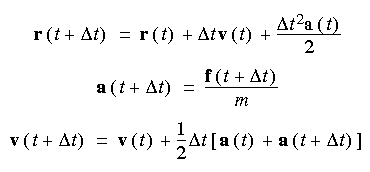
In order to calculate the forces of attraction and repulsion, f, between the molecules, use the formula for the Lennard-Jones force (FLJ). Use the parameters corresponding to water molecules in the force equation.
Use 1 unit for mass, m.
You program will be called as <prog_name> <input_file>.
Following is the format of the input file:
<number of particles, n>
<time interval, deltaT>
<n number of (x, y) positions, r>
< n number of (x, y) velocities, v>
<n number of (x, y) accelerations, r>
Your task is to load balance the code taking into account the load on the systems. During demo, after I start the run, I will be introducing artificial load during execution. You must build a load balancing strategy within your code to take care of this. Besides the demo, your task is compare the execution times of the load balanced and the non-load balanced versions for different conditions (problem sizes, different introduction of loads etc.) and write a small report based on the results, comparison, analysis. Remember that since the application load also frequently changes, it is not clear if your load balancing techniques will always yield benefits.
You can use few machines from the students' lab for this purpose.